SIB / SciLifeLab Autumn School Single Cell Analysis - October 2019
Section outline
-
Joint SIB / SciLifeLab Autumn School Single Cell Analysis
13 - 18 October 2019 - Leysin, Switzerland
This page is addressed to registered participants. To access course description and application form, please click here.
For any assistance, please contact training@sib.swiss.

-
Location: Hotel Central Residence & Spa, Leysin, Switzerland
Travel:
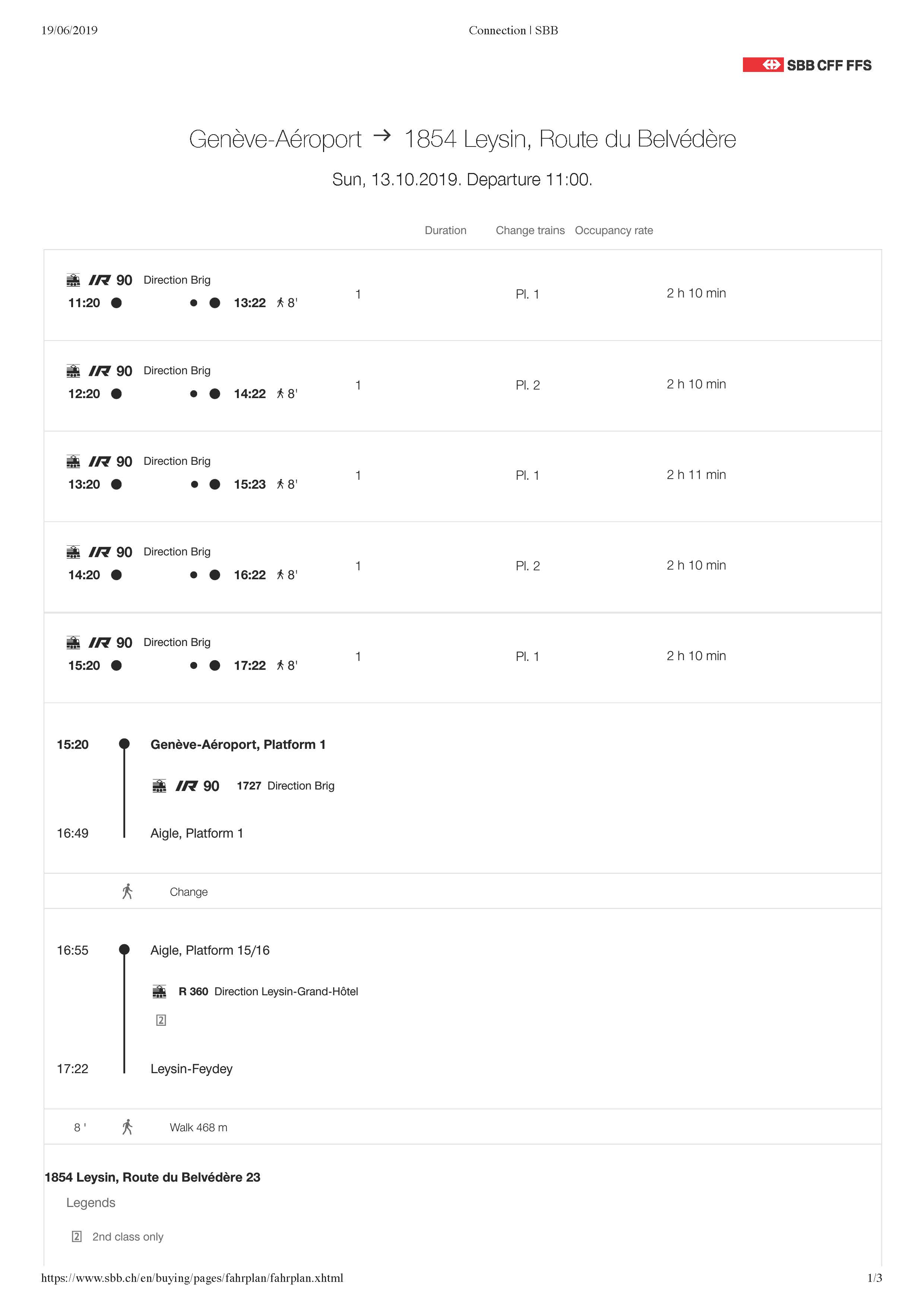
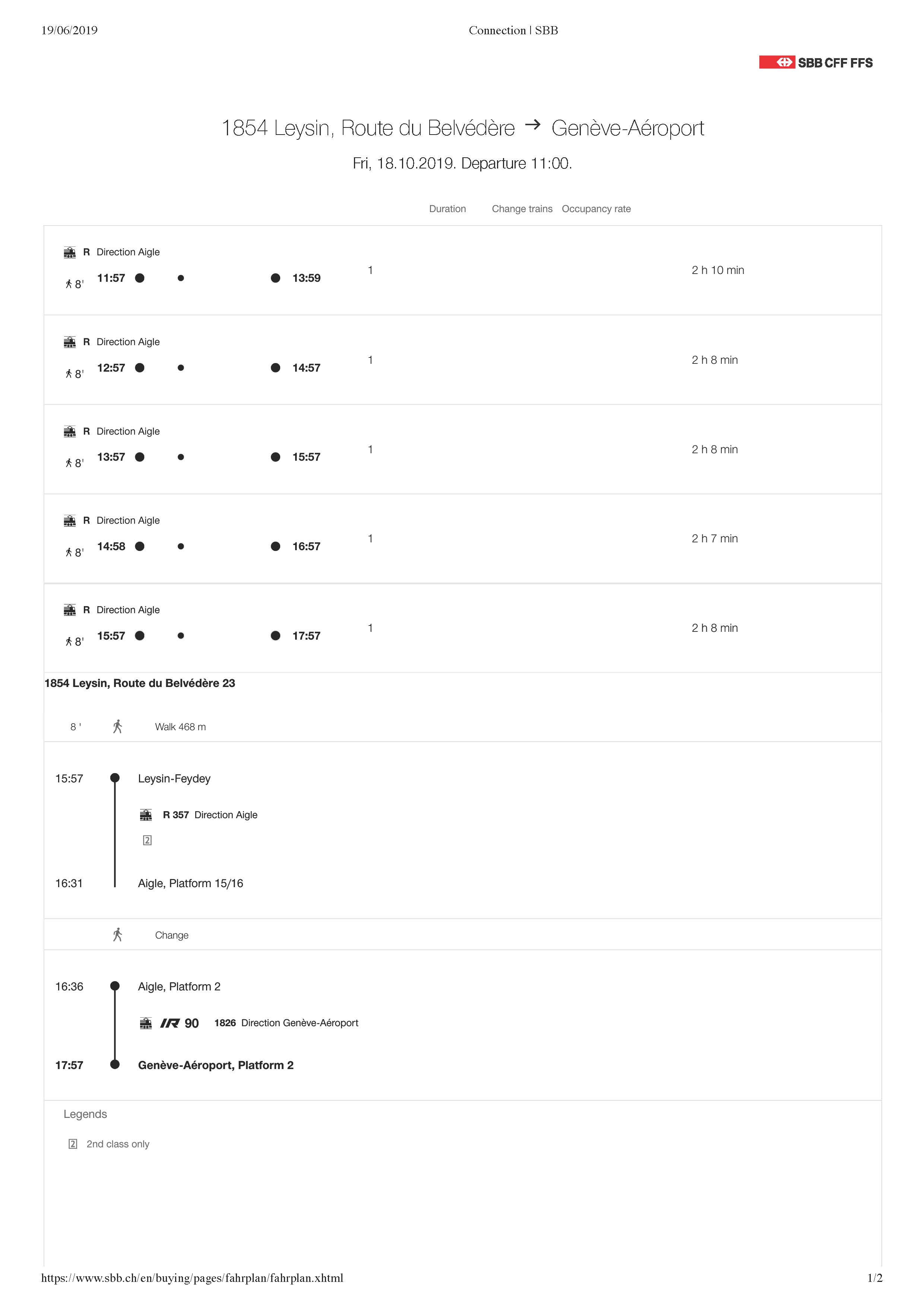
By plane: arrival at Geneva-Airport. Trains 1x / hour every xx:20 direction Brig, change at Aigle. Travel time 2h10. Detailed timetable below.
By train only: wherever your starting point, the last (small) train will be from Aigle to Leysin-Feydey, then a few minutes walk
Detailed timetable on https://www.sbb.ch/en/home.html
By car: Hotel address: route du Belvédère 20, CH-1854 Leysin
- Social activity & free time: if weather is not too bad we will go for hiking on Wednesday afternoon, so please bring good walking shoes and warm clothes. The hotel is equipped with a small spa, don't forget your swimsuit.
-
Usual schedule:
Start: 9:00
End: between 17:00 and 18:00
Lunch break: around 1h30
Coffee breaks: usually 2x per day
Dinner: around 20:00
Free time / swimming pool: 18-20:00 (open from 8 to 21)Sunday
Until 18h: arrival of the participants, check-in
18:45 Welcome drink and dinner
Monday: Transcriptomics
9:00 - 10:00 Introductory lecture: Vincent Gardeux, Laboratory of Systems and Genetics, EPFL / SIB, Lausanne, Switzerland.
10:00 - 11:00 Quantification, QC & Normalization: Davide Risso, Department of Statistical Sciences, University of Padova, Italy.
11:00 - 11:30 Coffee break
11:30 - 12:30 Dimensionality reduction: Paulo Czarnewski, NBIS, Stockholm University, SciLifeLab, Stockholm, Sweden.
12:30 - 14:00 Lunch break
14:00 - 18:00 hands-on including a coffee break (David + Paulo)
18:00 - 20:00 free time / spa
20:00 Dinner
Tuesday: Transcriptomics
9:00 - 10:00 Batch correction: Panagiotis Papasaikas, Computational Biology Group, Friedrich Miescher Institute for Biomedical Research / SIB, Basel. Switzerland.
10:00 - 11:00 Clustering - methods overview: Charlotte Soneson, Computational Biology Group, Friedrich Miescher Institute for Biomedical Research / SIB, Basel, Switzerland.
11:00 - 11:30 Coffee break
11:30 - 12:30 Cell fate mapping and trajectories: Wouter Saelens, VIB-UGent Center for Inflammation Research, Gent, Belgium.
12:30 - 14:00 Lunch break
14:00 - 18:00 hands-on including a coffee break (Panagiotis, Charlotte, Wouter)
18:00 - 20:00 free time / spa
20:00: special dinner - Cheese fondue
Wednesday: Transcriptomics
9:00 - Differential expression: Charlotte Soneson, Computational Biology, Friedrich Miescher Institute for Biomedical Research / SIB, Basel, Switzerland + short hands-on session.
~12:30 Lunch break
Afternoon: Team activity
18:30 - 19:30 Keynote lecture: Alejandro Sifrim, Laboratory of Reproductive Genomics, KU Leuven, Belgium.
20:00 Dinner
Thursday: Proteomics
9:00 - 10:30 Transcriptome + proteome: Johan Reimegård, Uppsala University, SciLifeLab, Uppsala, Sweden.
10:30 - 11:00 Coffee break
11:00 - 12:30 Differential abundance and differential state analysis of single cell cytometry data, Mark Robinson & Helena Crowell, Statistical Genomics, University of Zurich / SIB, Zurich, Switzerland.
12:30 - 14:00 Lunch break
14:00-15:45 hands-on part 1
15:45-16:15 coffee break
16:15:18:00 hands-on part 2 (Mark, Helena, Johan)
18:45 Dinner
<20:00 - 22:00 Special late opening of the spa
Friday: Other omics and integration
9:00 - 10:00 Data integration methods: Sebastien Smallwood, Friedrich Miescher Institute for Biomedical Research, Basel, Switzerland.
10:00 - 11:00 Spatial mapping of scRNAseq: Lars Borm, Department of Medical Biochemistry and Biophysics, Karolinska Institutet, Stockholm, Sweden.
11:00 - 11:30 Coffee break
11:30 - 12:30 Lineage tracing and scRNA: Maria Florescu, Hubrecht Institute, Developmental Biology and Stem Cell Research, Utrecht, Netherlands.
12:30 - 14:00 Lunch break
14:00 - 14:20 Conclusion and wrap up
14:20 End of the event
14:58 Train from Leysin-Feydey station
