Introduction to protein structure data representation and visualization
Section outline
-
Protein structure information is represented in data formats specifying the coordinates and properties of individual atoms. A variety of different visualization and abstraction methods has been developed to allow a human eye to comprehend molecular properties of complex macromolecules.
Depending on which area you work in, certain preferences for representing molecular information are used interchangeably for Alanine:
A
Ala
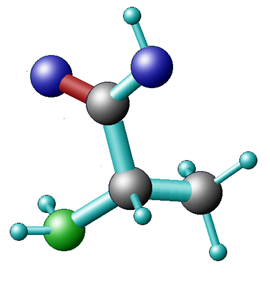
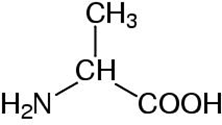
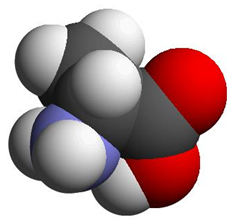
a potential computational chemists view chemical structure formula CPK representation The International Chemical Identifier (InChI) is used to unambiguously represent chemical entities in a string format:
InChI=1S/C3H7NO2/c1-2(4)3(5)6/h2H,4H2,1H3,(H,5,6)/t2-/m0/s1
The InChI notation is used for ligands, not for entire proteins.
Another very common notation is the widely used Simplified molecular-input line-entry system (SMILES) format (also for Alanine), although that is known to have short-comings concerning stereochemistry:
C[C@@H](C(=O)O)NThe SMILES notation is also used for ligands, not for entire proteins.
Likewise structural biologists have (ab-)used the ProteinDataBank (PDB) format intensively from the beginning. The format is column-based and thus ill-suited to describe entries with more than e.g. 99999 atoms or 9999 residues (see below for a superceding format PDBx/mmcif). There is also a the new mmtf format, which is beyond the scope of this course.
ATOM 263 N ALA A 35 1.429 34.959 -16.825 1.00 35.48 N
ATOM 264 CA ALA A 35 0.523 34.398 -17.829 1.00 35.10 C
ATOM 265 C ALA A 35 -0.724 33.878 -17.157 1.00 33.88 C
ATOM 266 O ALA A 35 -1.850 34.138 -17.600 1.00 33.13 O
ATOM 267 CB ALA A 35 1.209 33.268 -18.594 1.00 33.84 C
Recently the PDBX/mmcif format has been chosen as the main representation for all molecular data.
The format is easily extensible for more complex biological structures. The mmcif structure for Alanine:data_ALA # _chem_comp.id ALA _chem_comp.name ALANINE _chem_comp.type "L-PEPTIDE LINKING" _chem_comp.pdbx_type ATOMP _chem_comp.formula "C3 H7 N O2" _chem_comp.mon_nstd_parent_comp_id ? _chem_comp.pdbx_synonyms ? _chem_comp.pdbx_formal_charge 0 _chem_comp.pdbx_initial_date 1999-07-08 _chem_comp.pdbx_modified_date 2011-06-04 _chem_comp.pdbx_ambiguous_flag N _chem_comp.pdbx_release_status REL _chem_comp.pdbx_replaced_by ? _chem_comp.pdbx_replaces ? _chem_comp.formula_weight 89.093 _chem_comp.one_letter_code A _chem_comp.three_letter_code ALA _chem_comp.pdbx_model_coordinates_details ? _chem_comp.pdbx_model_coordinates_missing_flag N _chem_comp.pdbx_ideal_coordinates_details ? _chem_comp.pdbx_ideal_coordinates_missing_flag N _chem_comp.pdbx_model_coordinates_db_code ? _chem_comp.pdbx_subcomponent_list ? _chem_comp.pdbx_processing_site RCSB # loop_ _chem_comp_atom.comp_id _chem_comp_atom.atom_id _chem_comp_atom.alt_atom_id _chem_comp_atom.type_symbol _chem_comp_atom.charge _chem_comp_atom.pdbx_align _chem_comp_atom.pdbx_aromatic_flag _chem_comp_atom.pdbx_leaving_atom_flag _chem_comp_atom.pdbx_stereo_config _chem_comp_atom.model_Cartn_x _chem_comp_atom.model_Cartn_y _chem_comp_atom.model_Cartn_z _chem_comp_atom.pdbx_model_Cartn_x_ideal _chem_comp_atom.pdbx_model_Cartn_y_ideal _chem_comp_atom.pdbx_model_Cartn_z_ideal _chem_comp_atom.pdbx_component_atom_id _chem_comp_atom.pdbx_component_comp_id _chem_comp_atom.pdbx_ordinal ALA N N N 0 1 N N N 2.281 26.213 12.804 -0.966 0.493 1.500 N ALA 1 ALA CA CA C 0 1 N N S 1.169 26.942 13.411 0.257 0.418 0.692 CA ALA 2 ALA C C C 0 1 N N N 1.539 28.344 13.874 -0.094 0.017 -0.716 C ALA 3 ALA O O O 0 1 N N N 2.709 28.647 14.114 -1.056 -0.682 -0.923 O ALA 4 ALA CB CB C 0 1 N N N 0.601 26.143 14.574 1.204 -0.620 1.296 CB ALA 5 ALA OXT OXT O 0 1 N Y N 0.523 29.194 13.997 0.661 0.439 -1.742 OXT ALA 6 ALA H H H 0 1 N N N 2.033 25.273 12.493 -1.383 -0.425 1.482 H ALA 7 ALA H2 HN2 H 0 1 N Y N 3.080 26.184 13.436 -0.676 0.661 2.452 H2 ALA 8 ALA HA HA H 0 1 N N N 0.399 27.067 12.613 0.746 1.392 0.682 HA ALA 9 ALA HB1 1HB H 0 1 N N N -0.247 26.699 15.037 1.459 -0.330 2.316 HB1 ALA 10 ALA HB2 2HB H 0 1 N N N 0.308 25.110 14.270 0.715 -1.594 1.307 HB2 ALA 11 ALA HB3 3HB H 0 1 N N N 1.384 25.876 15.321 2.113 -0.676 0.697 HB3 ALA 12 ALA HXT HXT H 0 1 N Y N 0.753 30.069 14.286 0.435 0.182 -2.647 HXT ALA 13 # loop_ _chem_comp_bond.comp_id _chem_comp_bond.atom_id_1 _chem_comp_bond.atom_id_2 _chem_comp_bond.value_order _chem_comp_bond.pdbx_aromatic_flag _chem_comp_bond.pdbx_stereo_config _chem_comp_bond.pdbx_ordinal ALA N CA SING N N 1 ALA N H SING N N 2 ALA N H2 SING N N 3 ALA CA C SING N N 4 ALA CA CB SING N N 5 ALA CA HA SING N N 6 ALA C O DOUB N N 7 ALA C OXT SING N N 8 ALA CB HB1 SING N N 9 ALA CB HB2 SING N N 10 ALA CB HB3 SING N N 11 ALA OXT HXT SING N N 12 # loop_ _pdbx_chem_comp_descriptor.comp_id _pdbx_chem_comp_descriptor.type _pdbx_chem_comp_descriptor.program _pdbx_chem_comp_descriptor.program_version _pdbx_chem_comp_descriptor.descriptor ALA SMILES ACDLabs 10.04 "O=C(O)C(N)C" ALA SMILES_CANONICAL CACTVS 3.341 "C[C@H](N)C(O)=O" ALA SMILES CACTVS 3.341 "C[CH](N)C(O)=O" ALA SMILES_CANONICAL "OpenEye OEToolkits" 1.5.0 "C[C@@H](C(=O)O)N" ALA SMILES "OpenEye OEToolkits" 1.5.0 "CC(C(=O)O)N" ALA InChI InChI 1.03 "InChI=1S/C3H7NO2/c1-2(4)3(5)6/h2H,4H2,1H3,(H,5,6)/t2-/m0/s1" ALA InChIKey InChI 1.03 QNAYBMKLOCPYGJ-REOHCLBHSA-N # loop_ _pdbx_chem_comp_identifier.comp_id _pdbx_chem_comp_identifier.type _pdbx_chem_comp_identifier.program _pdbx_chem_comp_identifier.program_version _pdbx_chem_comp_identifier.identifier ALA "SYSTEMATIC NAME" ACDLabs 10.04 L-alanine ALA "SYSTEMATIC NAME" "OpenEye OEToolkits" 1.5.0 "(2S)-2-aminopropanoic acid" # loop_ _pdbx_chem_comp_audit.comp_id _pdbx_chem_comp_audit.action_type _pdbx_chem_comp_audit.date _pdbx_chem_comp_audit.processing_site ALA "Create component" 1999-07-08 RCSB ALA "Modify descriptor" 2011-06-04 RCSB #
